The idea that SARS-CoV-2 evolved from (is descended from) RaTG13 (or very similar virus) is rejected by the genome data. There are >1100 nucleotide differences - but it is not as simple as positing an order of magnitude elevated rate of evolution…
Mon Oct 11 10:17:02 +0000 2021
Replying to @arambaut
Half of these mutation would need to be reverting the changes that have occurred on lineage leading to RaTG13 since its last common ancestor with SARS-CoV-2. https://doi.org/10.1038/s41564-020-0771-4
Mon Oct 11 10:20:45 +0000 2021
Replying to @arambaut
Many of these mutations are synonymous and thus one cannot assert that these reversions would be due to selection.
Mon Oct 11 10:22:53 +0000 2021
Replying to @arambaut
There is also no evidence that the lineage leading SARS-CoV-2 has undergone any recombination since its last common ancestor with RaTG13 (or the recent Laotian bat viruses). The lineage leading to RaTG13 did under go recombination acquiring a different variable loop in the RBD
Mon Oct 11 10:25:46 +0000 2021
Replying to @arambaut
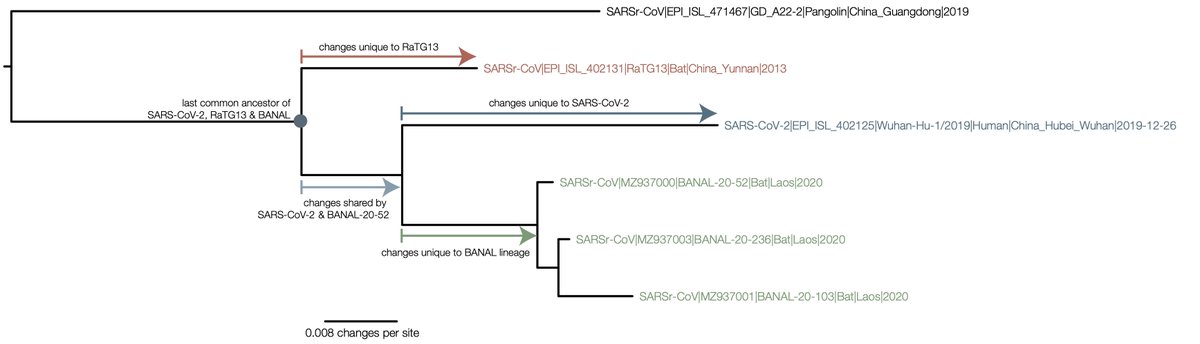
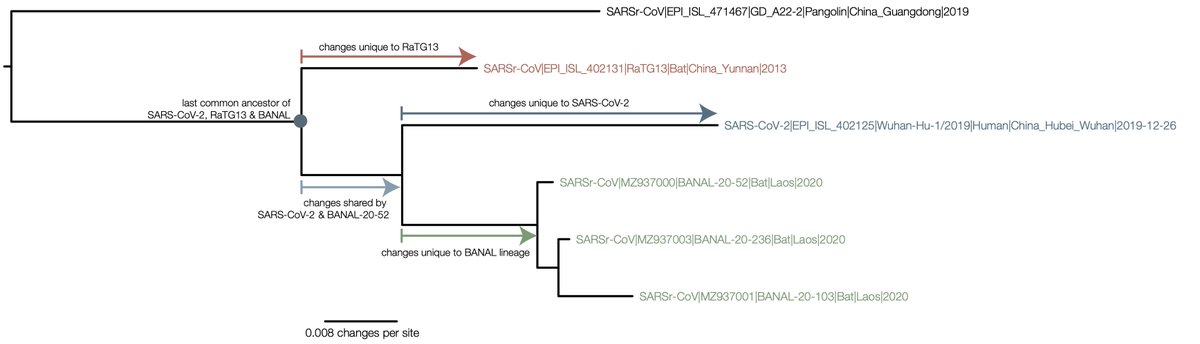
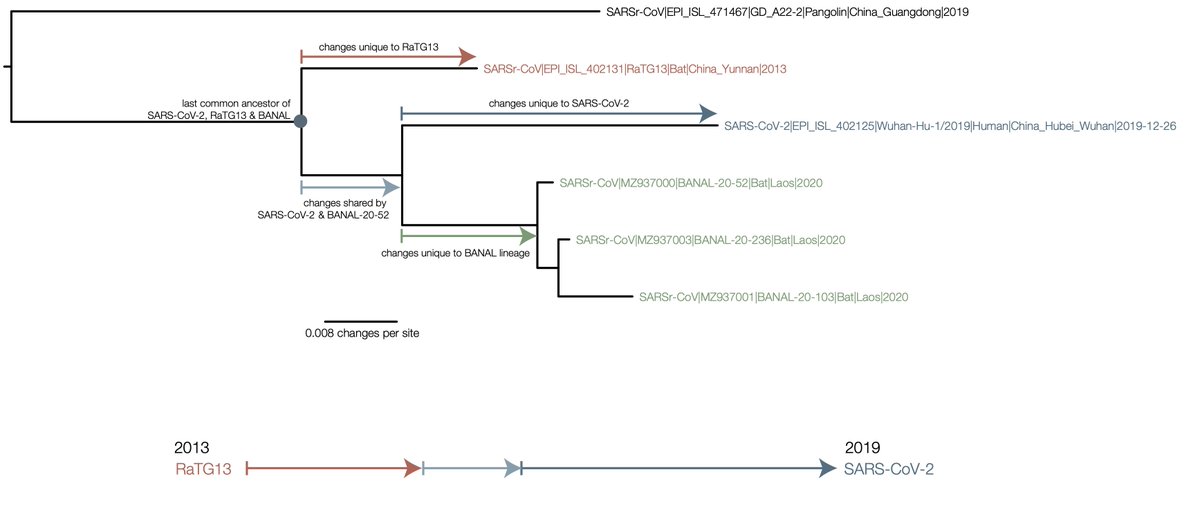
To clarify this further, here is a diagram of the phylogeny (this is for the S2 domain of spike but the same argument applies to the rest if the trees in https://doi.org/10.1038/s41564-020-0771-4
Mon Oct 11 10:57:10 +0000 2021
Replying to @arambaut
To go from RaTG13 to SARS-CoV-2 you would need to undo all of the mutations that are inferred on the red branch, then acquire the ones on the light blue branch (shared with the Laotian BANAL genomes) and finally the dark blue (unique to SARS-CoV-2).
Mon Oct 11 11:00:44 +0000 2021
Replying to @arambaut
The other way of putting it is the direct evolution of RaTG13 to SARS-CoV-2 would have to give the signal of the phylogenetic tree (i.e., the figure at the bottom of the image).
Mon Oct 11 11:07:39 +0000 2021